Introduction
This vignette demonstrates a full ezTrack workflow: importing tracking data with non-standard column names, cleaning and standardizing it, converting it to a spatial object, generating summary tables, visualizing fix rates and latitude over time, and plotting tracks and home ranges on an interactive map.
1. Import and format tracking data
We pass a dataset of GPS-tagged migratory shorebirds to
ez_track(). If column names aren’t provided,
ez_track() will automatically attempt to guess which
columns correspond to ID, timestamp, longitude, and latitude.
data(godwit_tracks)
head(godwit_tracks)
#> individual.local.identifier timestamp location.long location.lat
#> 3935 animal1 2024-12-31 16:00:00 -15.84644 16.33822
#> 3936 animal1 2024-12-31 20:00:00 -15.82790 16.38281
#> 3937 animal1 2024-12-31 22:00:00 -15.82829 16.38226
#> 3938 animal1 2025-01-01 00:00:00 -15.82715 16.38327
#> 3939 animal1 2025-01-01 04:00:00 -15.82761 16.38304
#> 3940 animal1 2025-01-01 10:00:00 -15.84629 16.32770
tracks <- ez_track(
data = godwit_tracks
)
#> Detected columns - id: individual.local.identifier, timestamp: timestamp, x: location.long, y: location.lat
#> Removed 852 row(s) with missing or duplicate values.If automatic column guessing fails or guesses incorrectly, you can manually specify which columns represent the ID, timestamp, longitude, and latitude.
tracks_manual <- ez_track(
data = godwit_tracks,
id_col = 'individual.local.identifier',
time_col = 'timestamp',
x_col = 'location.long',
y_col = 'location.lat',
)
#> Detected columns - id: individual.local.identifier, timestamp: timestamp, x: location.long, y: location.lat
#> Removed 852 row(s) with missing or duplicate values.You can also subsample the dataset using the subsample()
argument, which limits the number of location fixes per time unit. Use a
simple string format such as “1 per hour” or “2 per day” to control the
sampling frequency.
tracks_subsampled <- ez_track(
data = godwit_tracks,
subsample = "2 per hour"
)
#> Detected columns - id: individual.local.identifier, timestamp: timestamp, x: location.long, y: location.lat
#> Removed 852 row(s) with missing or duplicate values.
#> Subsampled to 2 fix(es) per hour2. Tracking data summary reports
Summary reports can be computed for each individual using
ez_summary(). For easy use in reports or presentations,
HTML reports can be generated using report = TRUE.
Summaries can also be filtered by date using the start_date
and end_date arguments.
The following summary statistics are returned for each unique id:
- n_fixes: Number of location records
- first_location: Timestamp of the first recorded location
- last_location: Timestamp of the last recorded location
- tracking_duration_days: Duration between first and last fix (in
days)
- fixes_per_day: Average number of fixes per day
- median_interval_hours: Median interval between fixes (in
hours)
- max_time_gap_days: Longest time gap between consecutive fixes (in
days)
- distance_km: Total distance traveled (in kilometers), calculated
using the Haversine formula
- avg_speed_kmh: Average speed (km/h), computed as distance divided by tracking duration in hours
summary_table <- ez_summary(tracks)
summary_table
#> id n_fixes first_location last_location
#> 1 animal1 1095 2024-12-31 16:00:00 2025-04-25 14:00:00
#> 2 animal2 1153 2024-12-31 16:00:00 2025-04-27 22:00:00
#> 3 animal3 1022 2024-12-31 16:00:00 2025-04-23 16:00:00
#> 4 animal4 214 2024-12-31 16:00:00 2025-04-27 16:00:00
#> 5 animal5 223 2024-12-31 16:00:00 2025-04-27 16:00:00
#> 6 animal6 551 2024-12-31 16:00:00 2025-04-27 16:00:00
#> tracking_duration_days fixes_per_day median_interval_hours max_time_gap_days
#> 1 114.92 9.53 2 0.33
#> 2 117.25 9.83 2 1.00
#> 3 113.00 9.04 2 1.00
#> 4 117.00 1.83 6 10.00
#> 5 117.00 1.91 6 13.38
#> 6 117.00 4.71 4 1.00
#> distance_km avg_speed_kmh
#> 1 5649.19 2.05
#> 2 5495.39 1.95
#> 3 6370.80 2.35
#> 4 2414.07 0.86
#> 5 2372.16 0.84
#> 6 5438.92 1.943. Visualize latitude over time
ez_latitude_plot() can be used to visualize latitudinal
movement (north-south) over time for each tracked animal.
ez_latitude_plot(tracks)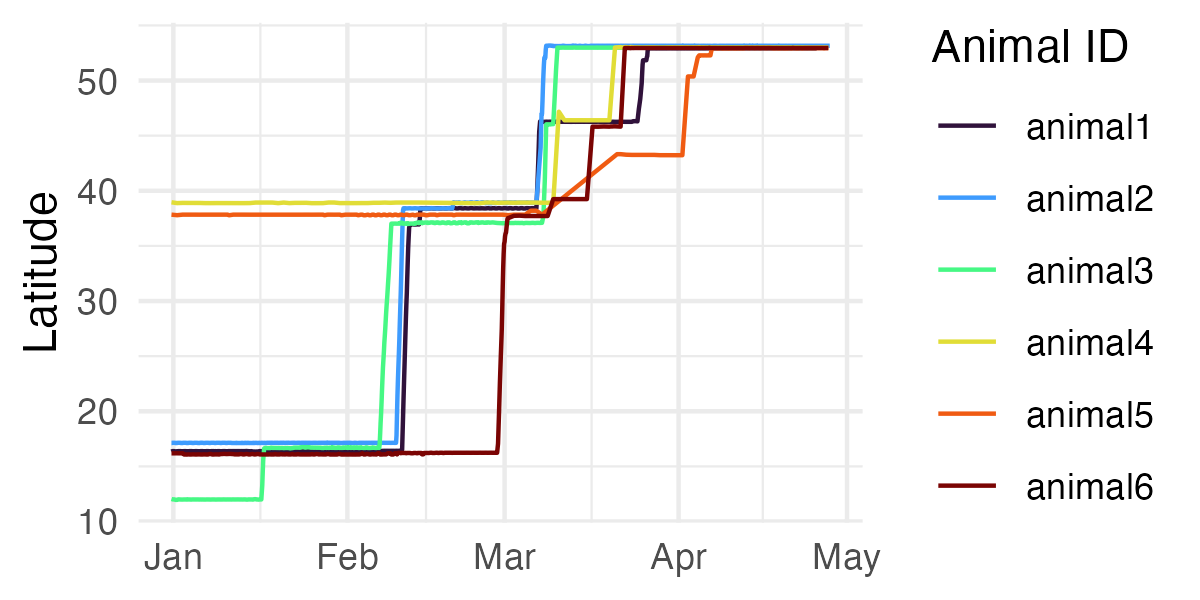
The plot supports:
Optional faceting by individual
Date axis customization
Date range filtering
ggplot2parameters for further customization.
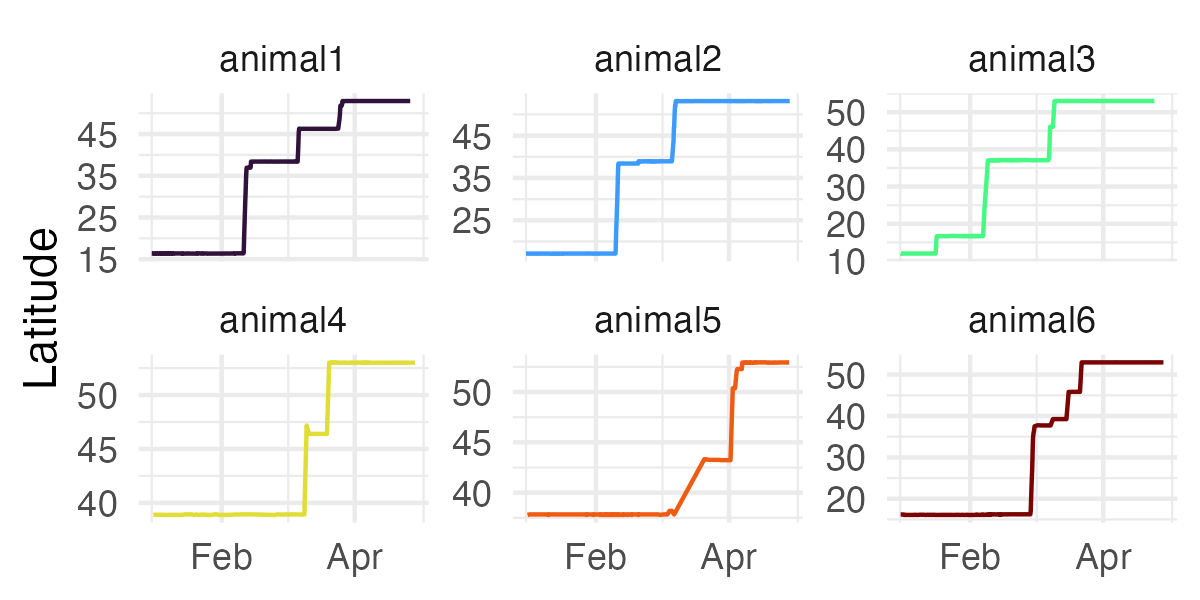
ez_latitude_plot(
tracks,
facet = TRUE,
start_date = "2025-01-01",
end_date = "2025-04-28",
date_breaks = "2 months",
date_format = "%b"
)
4. Visualize fix rate over time
ez_fix_rate_plot() can be used to visualize fix rates
over time for each tracked animal, using a tick mark to represent each
location fix.
ez_fix_rate_plot(tracks)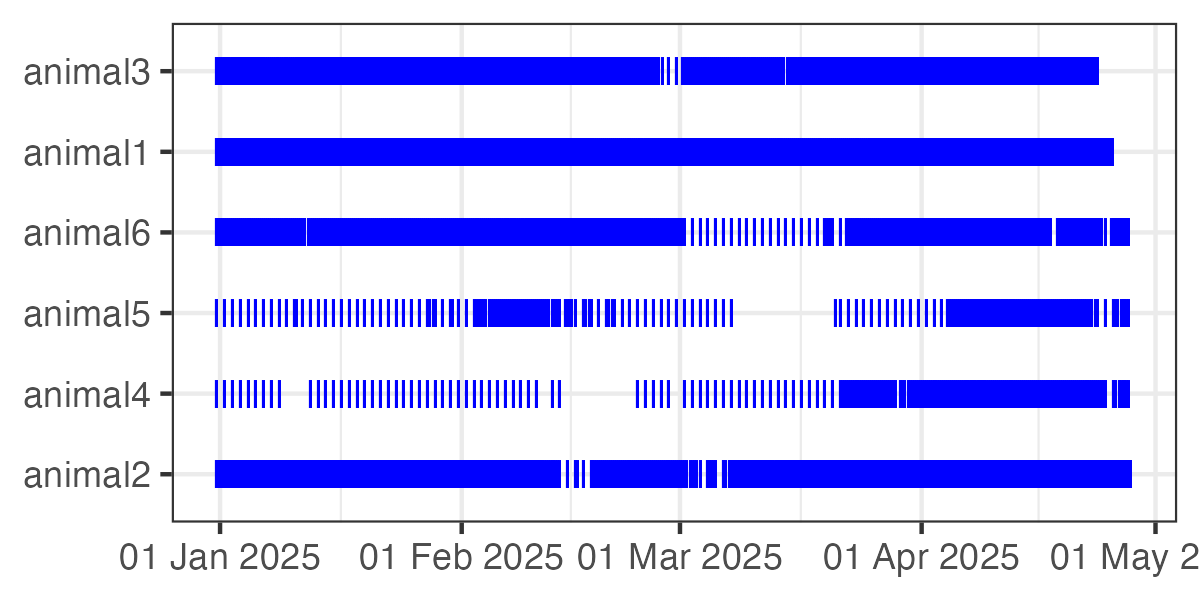
The plot supports:
Date axis customization
Date range filtering
ggplot2parameters for further customization.
5. Map tracks and home ranges
We can display tracks using an interactive Leaflet map with
ez_map() with a simple command.
ez_map(tracks)As well as with date range filters and a variety of style arguments.
ez_map(tracks,
start_date = "2025-04-10",
end_date = "2025-04-28",
point_color = "timestamp",
point_size = 2,
path_opacity = 0.3)6. Estimate home ranges
Individual or population-level Minimum Convex Polygons (MCP) or
Kernel Density Estimate (KDE) home ranges can be created with
ez_home_range().
home_ranges_mcp <- ez_home_range(tracks, method = 'mcp', level = 95, start_date = '2025-04-12')
#> Registered S3 methods overwritten by 'adehabitatMA':
#> method from
#> print.SpatialPixelsDataFrame sp
#> print.SpatialPixels spThe result is an sf object containing one home range
polygon per tracked individual or population (if
population = TRUE).
Home ranges can be visualized with ez_map.
ez_map(home_ranges = home_ranges_mcp)Home ranges can also be mapped alongside tracking data. In this example, we use the start_date argument to ensure the tracking data aligns temporally with the home range period.
ez_map(tracks, home_ranges = home_ranges_mcp, start_date = '2025-04-12')